This page was auto-generated from a Jupyter notebook: aniline/aniline_wf_0.1-10.1eV_orb33_A2.
Problems? Please raise any issues on Github.
ePSproc: Aniline Orb 33 (A2) ionization¶
electronic structure input: aniline_aug-cc-pVDZ_E_orbLT56.molden
ePS output file: aniline_wf_0.1-10.1eV_orb33_A2.inp.out
Web version: https://phockett.github.io/ePSdata/aniline/aniline_wf_0.1-10.1eV_orb33_A2.html
Dataset: https://zenodo.org/record/3612509
DOI (dataset): 10.5281/zenodo.3612509
Licensed under Creative Commons Attribution-NonCommercial-ShareAlike 4.0 (CC BY-NC-SA 4.0)
Job details¶
ePS aniline, batch aniline_wf_0.1-10.1eV, orbital orb33_A2
Aniline Orb 33 (A2) ionization, S1/LUMO test using B2 initial symmetry, B1 ion state, wavefn run on AntonJr
E=0.1:0.2:10.1 (51 points)
Thu Mar 7 17:45:08 EST 2019
Set-up¶
Load modules¶
[1]:
import sys
import os
import numpy as np
import epsproc as ep
from datetime import datetime as dt
timeString = dt.now()
* pyevtk not found, VTK export not available.
* plotly not found, plotly plots not available.
Load data¶
[2]:
# File path only, from env var DATAFILE
# dataPath = os.getcwd()
dataFile = os.environ.get('DATAFILE', '')
[3]:
jobInfo = ep.headerFileParse(dataFile)
molInfo = ep.molInfoParse(dataFile)
*** Job info from file header.
ePS aniline, batch aniline_wf_0.1-10.1eV, orbital orb33_A2
Aniline Orb 33 (A2) ionization, S1/LUMO test using B2 initial symmetry, B1 ion state, wavefn run on AntonJr
E=0.1:0.2:10.1 (51 points)
Thu Mar 7 17:45:08 EST 2019
*** Found orbitals
1 1 Ene = -15.5686 Spin =Alpha Occup = 2.000000
2 2 Ene = -11.2822 Spin =Alpha Occup = 2.000000
3 3 Ene = -11.2336 Spin =Alpha Occup = 2.000000
4 4 Ene = -11.2335 Spin =Alpha Occup = 2.000000
5 5 Ene = -11.2203 Spin =Alpha Occup = 2.000000
6 6 Ene = -11.2203 Spin =Alpha Occup = 2.000000
7 7 Ene = -11.2144 Spin =Alpha Occup = 2.000000
8 8 Ene = -1.2347 Spin =Alpha Occup = 2.000000
9 9 Ene = -1.1330 Spin =Alpha Occup = 2.000000
10 10 Ene = -1.0156 Spin =Alpha Occup = 2.000000
11 11 Ene = -0.9909 Spin =Alpha Occup = 2.000000
12 12 Ene = -0.8355 Spin =Alpha Occup = 2.000000
13 13 Ene = -0.8200 Spin =Alpha Occup = 2.000000
14 14 Ene = -0.7152 Spin =Alpha Occup = 2.000000
15 15 Ene = -0.7034 Spin =Alpha Occup = 2.000000
16 16 Ene = -0.6603 Spin =Alpha Occup = 2.000000
17 17 Ene = -0.6006 Spin =Alpha Occup = 2.000000
18 18 Ene = -0.5992 Spin =Alpha Occup = 2.000000
19 19 Ene = -0.5820 Spin =Alpha Occup = 2.000000
20 20 Ene = -0.5361 Spin =Alpha Occup = 2.000000
21 21 Ene = -0.5084 Spin =Alpha Occup = 2.000000
22 22 Ene = -0.4866 Spin =Alpha Occup = 2.000000
23 23 Ene = -0.4446 Spin =Alpha Occup = 2.000000
24 24 Ene = -0.3358 Spin =Alpha Occup = 2.000000
25 25 Ene = -0.2853 Spin =Alpha Occup = 2.000000
26 26 Ene = 0.0318 Spin =Alpha Occup = 0.000000
27 27 Ene = 0.0415 Spin =Alpha Occup = 0.000000
28 28 Ene = 0.0427 Spin =Alpha Occup = 0.000000
29 29 Ene = 0.0566 Spin =Alpha Occup = 0.000000
30 30 Ene = 0.0578 Spin =Alpha Occup = 0.000000
31 31 Ene = 0.0768 Spin =Alpha Occup = 0.000000
32 32 Ene = 0.0875 Spin =Alpha Occup = 0.000000
33 33 Ene = 0.1133 Spin =Alpha Occup = 0.000000
*** Found atoms
Z = 7 ZS = 7 r = 0.0000000000 0.0000000000 2.3057780000
Z = 6 ZS = 6 r = 0.0000000000 0.0000000000 0.9332450000
Z = 6 ZS = 6 r = 0.0000000000 1.1965340000 0.2222240000
Z = 6 ZS = 6 r = 0.0000000000 -1.1965340000 0.2222240000
Z = 6 ZS = 6 r = 0.0000000000 1.1882610000 -1.1569230000
Z = 6 ZS = 6 r = 0.0000000000 -1.1882610000 -1.1569230000
Z = 6 ZS = 6 r = 0.0000000000 0.0000000000 -1.8637550000
Z = 1 ZS = 1 r = 0.0000000000 2.1302880000 0.7533710000
Z = 1 ZS = 1 r = 0.0000000000 -2.1302880000 0.7533710000
Z = 1 ZS = 1 r = 0.0000000000 2.1240450000 -1.6836310000
Z = 1 ZS = 1 r = 0.0000000000 -2.1240450000 -1.6836310000
Z = 1 ZS = 1 r = 0.0000000000 0.0000000000 -2.9358510000
Z = 1 ZS = 1 r = 0.0000000000 -0.8462720000 2.8135300000
Z = 1 ZS = 1 r = 0.0000000000 0.8462720000 2.8135300000
[4]:
# Scan file(s) for various data types...
# For dir scan
# dataXS = ep.readMatEle(fileBase = dataPath, recordType = 'CrossSection')
# dataMatE = ep.readMatEle(fileBase = dataPath, recordType = 'DumpIdy')
# For single file
dataXS = ep.readMatEle(fileIn = dataFile, recordType = 'CrossSection')
dataMatE = ep.readMatEle(fileIn = dataFile, recordType = 'DumpIdy')
*** ePSproc readMatEle(): scanning files for CrossSection segments.
*** Scanning file(s)
['/home/paul/ePS_results/aniline/aniline_wf_0.1-10.1eV/aniline_wf_0.1-10.1eV_orb33_A2.inp.out']
*** Reading ePS output file: /home/paul/ePS_results/aniline/aniline_wf_0.1-10.1eV/aniline_wf_0.1-10.1eV_orb33_A2.inp.out
Expecting 51 energy points.
Expecting 3 symmetries.
Scanning CrossSection segments.
Expecting 4 CrossSection segments.
Found 4 CrossSection segments (sets of results).
Processed 4 sets of CrossSection file segments, (0 blank)
*** ePSproc readMatEle(): scanning files for DumpIdy segments.
*** Scanning file(s)
['/home/paul/ePS_results/aniline/aniline_wf_0.1-10.1eV/aniline_wf_0.1-10.1eV_orb33_A2.inp.out']
*** Reading ePS output file: /home/paul/ePS_results/aniline/aniline_wf_0.1-10.1eV/aniline_wf_0.1-10.1eV_orb33_A2.inp.out
Expecting 51 energy points.
Expecting 3 symmetries.
Scanning CrossSection segments.
Expecting 153 DumpIdy segments.
Found 153 dumpIdy segments (sets of matrix elements).
Processing segments to Xarrays...
Processed 153 sets of DumpIdy file segments, (0 blank)
Job & molecule info¶
[5]:
ep.jobSummary(jobInfo, molInfo);
*** Job summary data
ePS aniline, batch aniline_wf_0.1-10.1eV, orbital orb33_A2
Aniline Orb 33 (A2) ionization, S1/LUMO test using B2 initial symmetry, B1 ion state, wavefn run on AntonJr
E=0.1:0.2:10.1 (51 points)
Thu Mar 7 17:45:08 EST 2019
Electronic structure input: '/home/paul/ePS_stuff/aniline/electronic_structure/aniline_aug-cc-pVDZ_E_orbLT56.molden'
Initial state occ: [2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 1 0 0 0 0 0 0 0 1]
Final state occ: [2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 1 0 0 0 0 0 0 0 0]
IPot (input vertical IP, eV): 7.70
*** Additional orbital info (SymProd)
Ionizing orb: [0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1]
Ionizing orb sym: ['A2']
Orb energy (eV): [3.08305004]
Orb energy (H): [0.1133]
Orb energy (cm^-1): [24866.47573488]
Threshold wavelength (nm): 402.1478598985443
*** Molecular structure
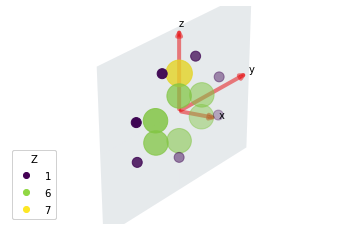
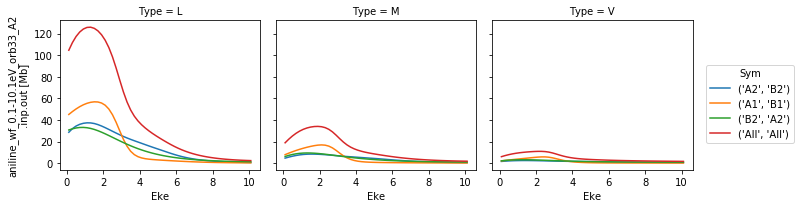
1-photon ePS Cross-Sections¶
Plot 1-photon cross-sections and \(beta_2\) parameters (for an unaligned ensemble) from ePS calculations. These are taken directly from the ePS output file, CrossSection segments. See the ePS manual, ``GetCro` command, for further details <https://www.chem.tamu.edu/rgroup/lucchese/ePolyScat.E3.manual/GetCro.html>`__.
Cross-sections by symmetry & type¶
Types correspond to:
‘L’: length gauge results.
‘V’: velocity gauge results.
‘M’: mixed gauge results.
Symmetries correspond to allowed ionizing transitions for the molecular point group (IRs typically corresponding to (x,y,z) polarization geometries), see the ePS manual for a list of symmetries. Symmetry All corresponds to the sum over all allowed sets of symmetries.
Cross-section units are MBarn.
[6]:
# Plot cross sections using Xarray functionality
# Set here to plot per file - should add some logic to combine files.
for data in dataXS:
daPlot = data.sel(XC='SIGMA')
daPlot.plot.line(x='Eke', col='Type')
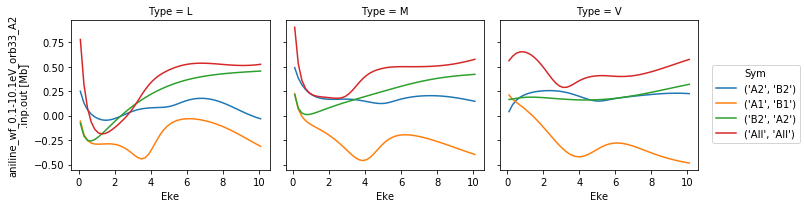
\(\beta_{2}\) by symmetry & type¶
Types & symmetries as per cross-sections. Normalized \(\beta_{2}\) paramters, dimensionless.
[7]:
# Repeat for betas
for data in dataXS:
daPlot = data.sel(XC='BETA')
daPlot.plot.line(x='Eke', col='Type')
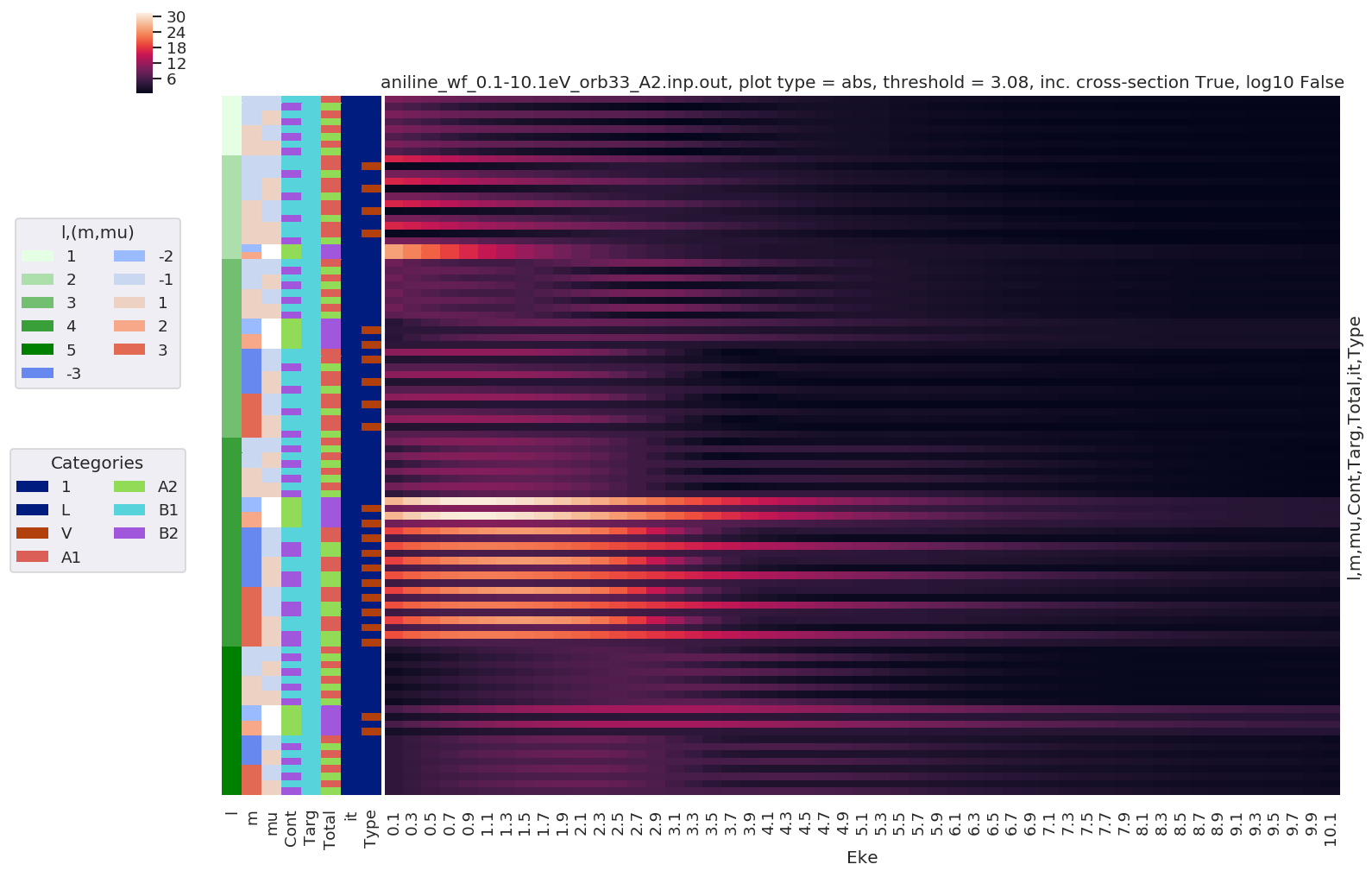
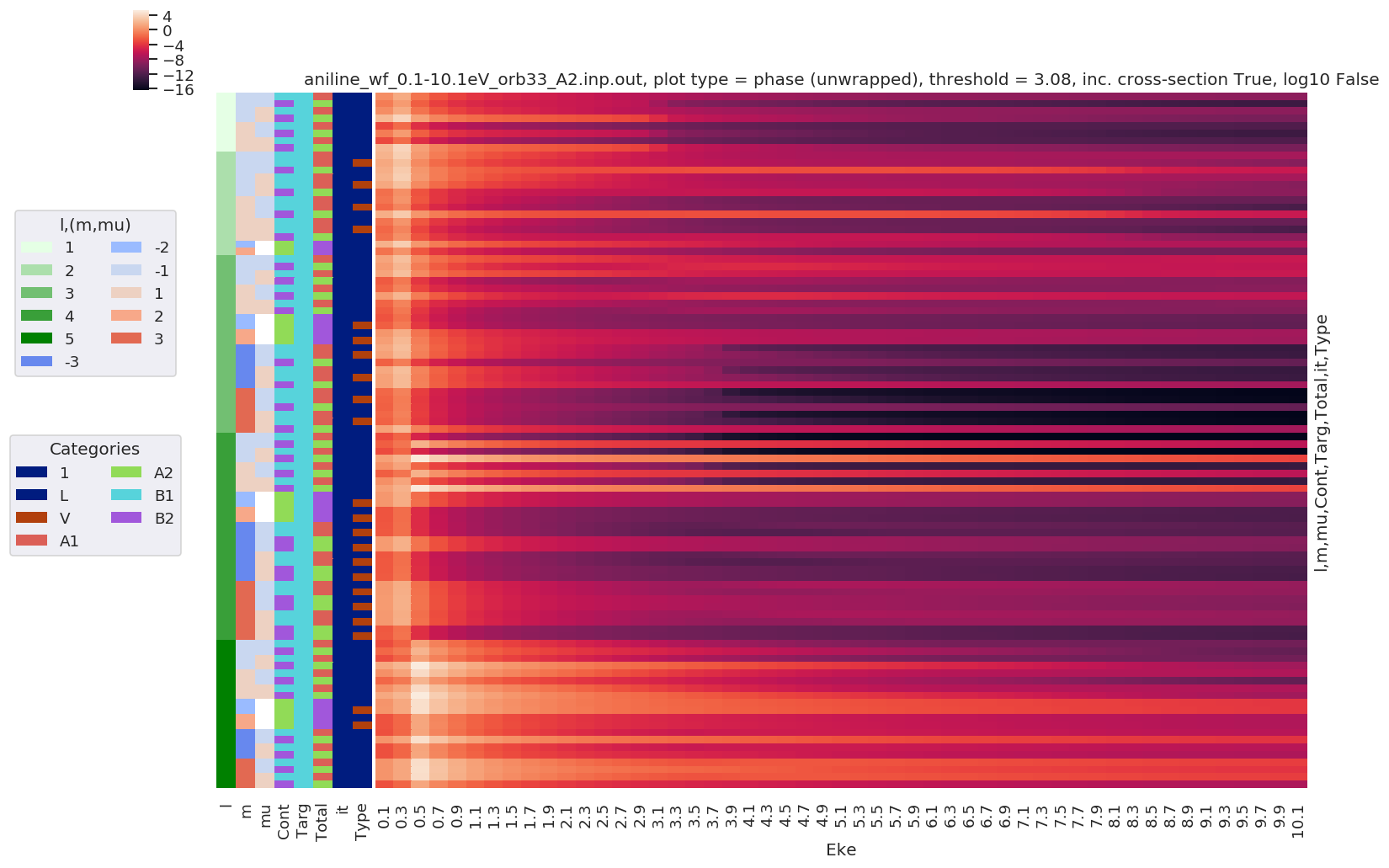
Dipole matrix elements¶
For 1-photon ionization. These are taken directly from ePS DumpIdy segments. See the ePS manual, ``DumpIdy` command, for further details <https://www.chem.tamu.edu/rgroup/lucchese/ePolyScat.E3.manual/DumpIdy.html>`__.
[8]:
# Set threshold for significance, only matrix elements with abs values > thres % will be plotted
thres = 0.1
[9]:
# Plot for each fie
for data in dataMatE:
# Plot with sensible defaults - all dims with lmPlot()
# Plot only values > theshold
daPlot, daPlotpd, legendList, gFig = ep.lmPlot(data, thres = thres, thresType = 'pc', figsize = (15,10))
# Plot phases, with unwrap
daPlot, daPlotpd, legendList, gFig = ep.lmPlot(data, thres = thres, thresType = 'pc', figsize = (15,10), pType='phaseUW')
/home/paul/anaconda3/envs/ePSproc-v1.2/lib/python3.7/site-packages/xarray/core/nputils.py:223: RuntimeWarning: All-NaN slice encountered
result = getattr(npmodule, name)(values, axis=axis, **kwargs)
Plotting data aniline_wf_0.1-10.1eV_orb33_A2.inp.out, pType=a, thres=3.0759236685324947, with Seaborn
/home/paul/anaconda3/envs/ePSproc-v1.2/lib/python3.7/site-packages/xarray/core/nputils.py:223: RuntimeWarning: All-NaN slice encountered
result = getattr(npmodule, name)(values, axis=axis, **kwargs)
/home/paul/anaconda3/envs/ePSproc-v1.2/lib/python3.7/site-packages/numpy/lib/function_base.py:1520: RuntimeWarning: invalid value encountered in greater
_nx.copyto(ddmod, pi, where=(ddmod == -pi) & (dd > 0))
/home/paul/anaconda3/envs/ePSproc-v1.2/lib/python3.7/site-packages/numpy/lib/function_base.py:1522: RuntimeWarning: invalid value encountered in less
_nx.copyto(ph_correct, 0, where=abs(dd) < discont)
Plotting data aniline_wf_0.1-10.1eV_orb33_A2.inp.out, pType=phaseUW, thres=3.0759236685324947, with Seaborn
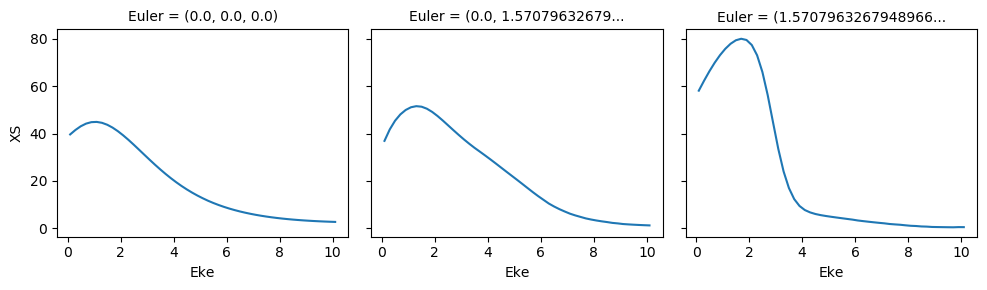
MFPADs¶
Calculated MF \(\beta\) parameters, using ePS dipole matrix elements. These are calculated by ep.mfblm(), as a function of energy and polarization geometry. See the ePSproc docs on ``ep.mfblm()` <https://epsproc.readthedocs.io/en/latest/modules/epsproc.MFBLM.html>`__ for further details, and this demo notebook.
[10]:
# Set pol geoms - these correspond to (z,x,y) in molecular frame (relative to principle/symmetry axis)
eAngs = ep.setPolGeoms()
[11]:
# Calculate for each fie & pol geom
# TODO - file logic, and parallelize
BLM = []
for data in dataMatE:
BLM.append(ep.mfblmEuler(data, selDims = {'Type':'L'}, eAngs = eAngs, thres = thres,
SFflag = True, verbose = 0)) # Run for all Eke, selected gauge only
[12]:
# Save BLM data - defaults to working dir and 'ep_timestamp' file
# TODO - testing for array/multiple file case
for data in BLM:
fileName = dataFile + '_BLM-L_' + timeString.strftime('%Y-%m-%d_%H-%M-%S')
ep.writeXarray(data, fileName = fileName)
['Written to h5netcdf format', '/home/paul/ePS_results/aniline/aniline_wf_0.1-10.1eV/aniline_wf_0.1-10.1eV_orb33_A2.inp.out_BLM-L_2020-01-14_08-50-18.nc']
/home/paul/anaconda3/envs/ePSproc-v1.2/lib/python3.7/site-packages/h5netcdf/core.py:481: H5pyDeprecationWarning: other_ds.dims.create_scale(ds, name) is deprecated. Use ds.make_scale(name) instead.
h5ds.dims.create_scale(h5ds, scale_name)
[13]:
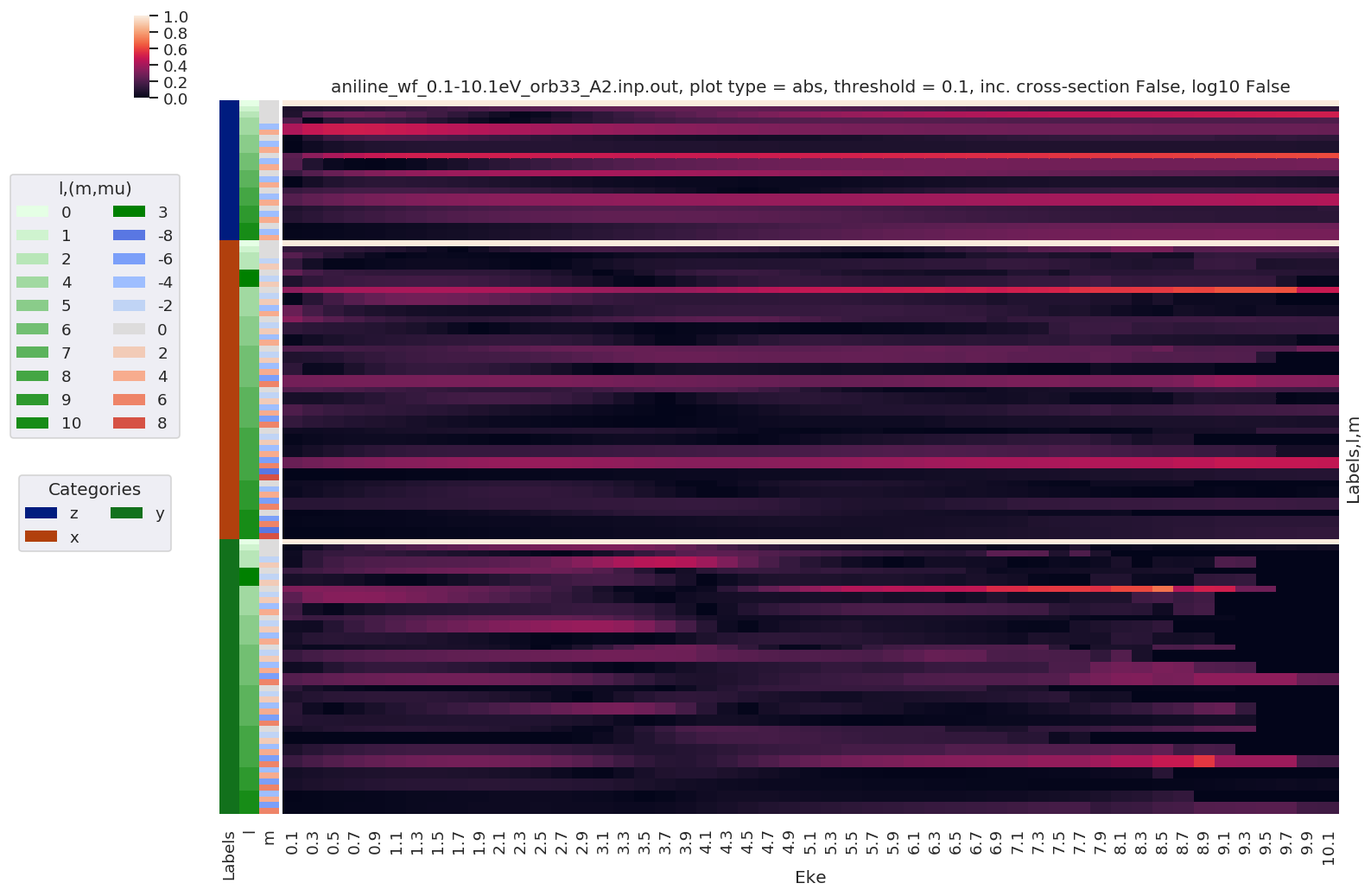
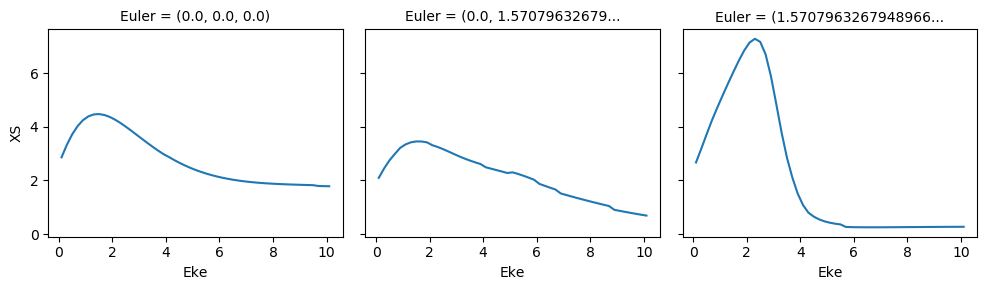
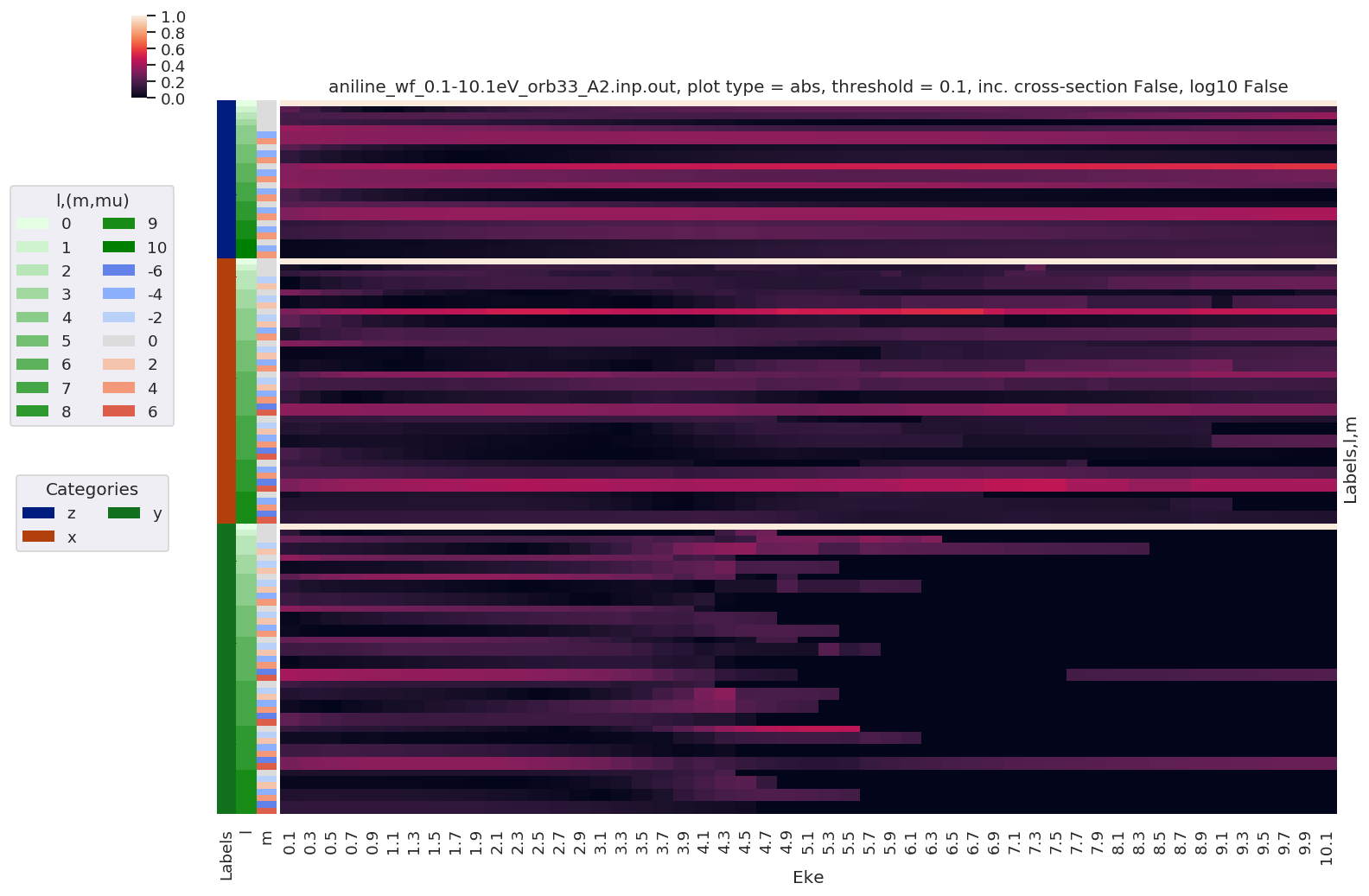
# Normalize and plot results
for BLMplot in BLM:
# Plot unnormalized B00 only, real part
# This is/should be in units of MBarn (TBC).
# BLMplot.where(np.abs(BLMplot) > thres, drop = True).real.squeeze().sel({'l':0, 'm':0}).plot.line(x='Eke', col='Euler');
BLMplot.XS.real.squeeze().plot.line(x='Eke', col='Euler');
# Plot values normalised by B00 - now set in calculation function
# Plot results with lmPlot(), ordering by Euler sets
# Version with (semi-manual) Euler grouping
daPlot, daPlotpd, legendList, gFig = ep.lmPlot(BLMplot.swap_dims({'Euler':'Labels'}), SFflag = False, eulerGroup = True,
thresType = 'pc', thres = thres,
plotDims = ('Labels','l','m'),
figsize = (15,10))
Plotting data aniline_wf_0.1-10.1eV_orb33_A2.inp.out, pType=a, thres=0.1, with Seaborn
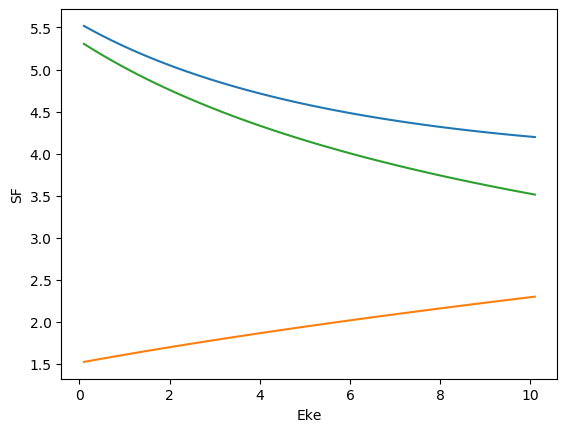
Error & consistency checks¶
[14]:
# Check SF values
for data in dataMatE:
# Plot values, single plot
data.SF.pipe(np.abs).plot.line(x='Eke')
data.SF.real.plot.line(x='Eke')
data.SF.imag.plot.line(x='Eke')
# Plot values, facet plot
# data.SF.pipe(np.abs).plot.line(x='Eke', col='Sym')
[15]:
# Compare calculated BLMs for L and V types (dafault above for L)
# Calculate for each fie & pol geom, and compare.
BLMv = []
BLMdiff = []
for n, data in enumerate(dataMatE):
BLMv.append(ep.mfblmEuler(data, selDims = {'Type':'V'}, eAngs = eAngs, thres = thres,
SFflag = True, verbose = 0)) # Run for all Eke, selected gauge only
BLMdiff.append(BLM[n] - BLMv[n])
BLMdiff[n]['dXS'] = BLM[n].XS - BLMv[n].XS # Set XS too, dropped in calc above
BLMdiff[n].attrs['dataType'] = 'matE'
[16]:
# Save BLM data - defaults to working dir and 'ep_timestamp' file
# TODO - testing for array/multiple file case
for data in BLMv:
fileName = dataFile + '_BLM-V_' + timeString.strftime('%Y-%m-%d_%H-%M-%S')
ep.writeXarray(data, fileName = fileName)
['Written to h5netcdf format', '/home/paul/ePS_results/aniline/aniline_wf_0.1-10.1eV/aniline_wf_0.1-10.1eV_orb33_A2.inp.out_BLM-V_2020-01-14_08-50-18.nc']
/home/paul/anaconda3/envs/ePSproc-v1.2/lib/python3.7/site-packages/h5netcdf/core.py:481: H5pyDeprecationWarning: other_ds.dims.create_scale(ds, name) is deprecated. Use ds.make_scale(name) instead.
h5ds.dims.create_scale(h5ds, scale_name)
[17]:
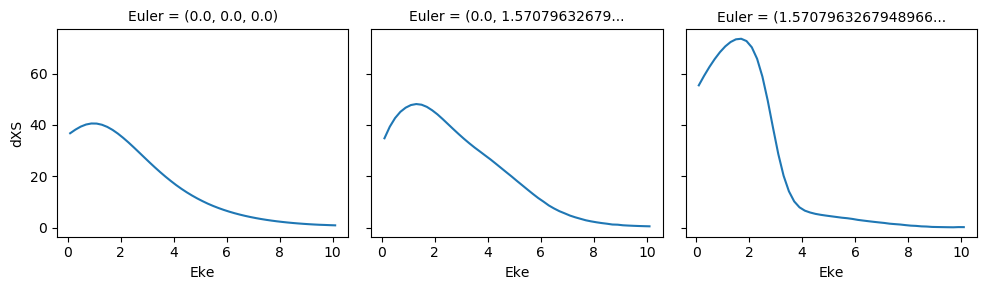
# Normalize and plot results
for BLMplot in BLMv:
# Plot unnormalized B00 only, real part
# This is/should be in units of MBarn (TBC).
# BLMplot.where(np.abs(BLMplot) > thres, drop = True).real.squeeze().sel({'l':0, 'm':0}).plot.line(x='Eke', col='Euler');
BLMplot.XS.real.squeeze().plot.line(x='Eke', col='Euler');
# Plot values normalised by B00 - now set in calculation function
# Plot results with lmPlot(), ordering by Euler sets
# Version with (semi-manual) Euler grouping
daPlot, daPlotpd, legendList, gFig = ep.lmPlot(BLMplot.swap_dims({'Euler':'Labels'}), SFflag = False, eulerGroup = True,
thresType = 'pc', thres = thres,
plotDims = ('Labels','l','m'),
figsize = (15,10))
Plotting data aniline_wf_0.1-10.1eV_orb33_A2.inp.out, pType=a, thres=0.1, with Seaborn
[18]:
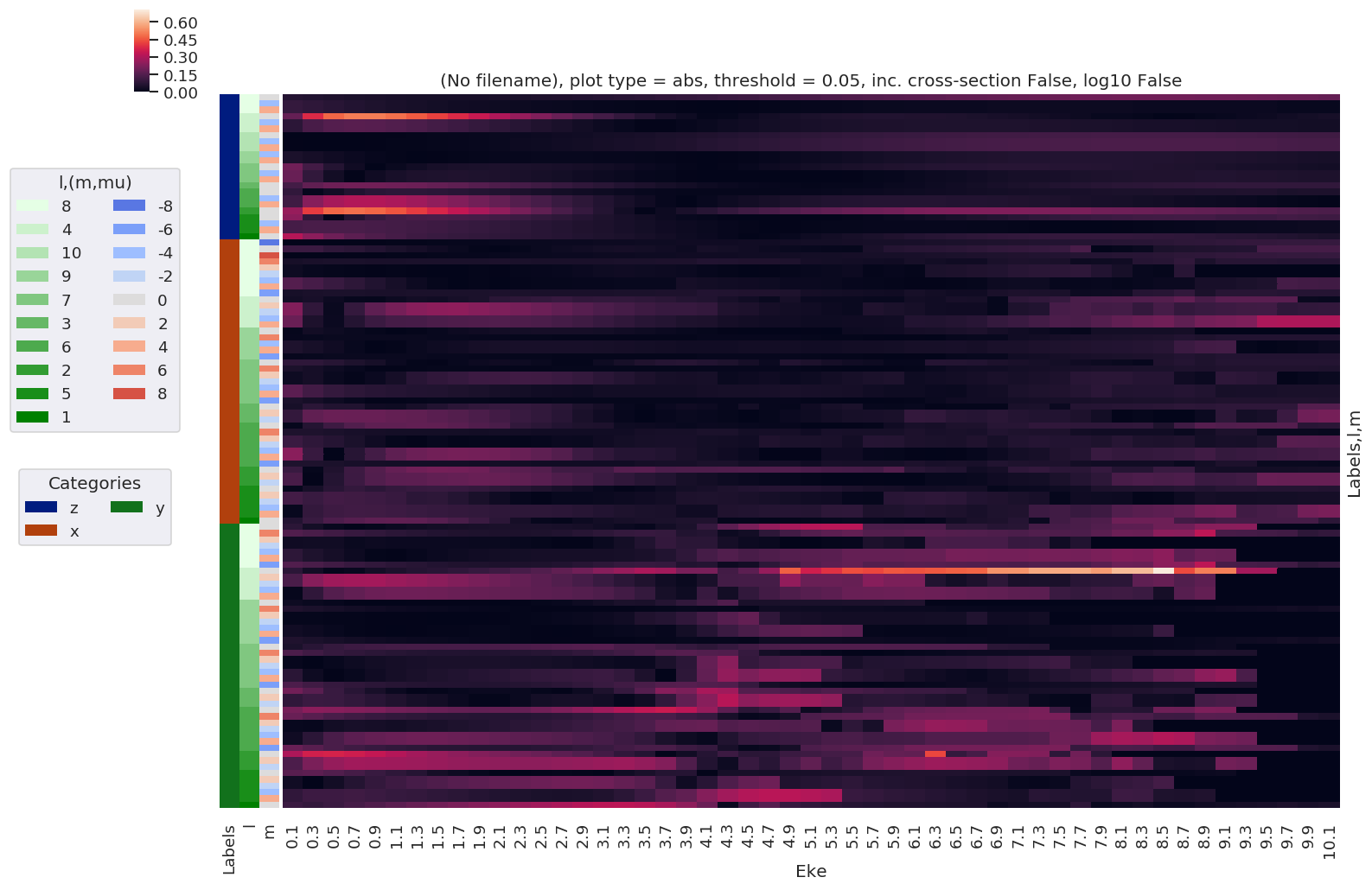
# Difference between 'L' and 'V' results
# NOTE - this currently drops XS
print('Differences, L vs. V gauge BLMs')
for BLMplot in BLMdiff:
maxDiff = BLMplot.max()
print(f'Max difference in BLMs (L-V): {0}', maxDiff.data)
if np.abs(maxDiff) > thres:
# Plot B00 only, real part
# BLMplot.where(np.abs(BLMplot) > thres, drop = True).real.squeeze().sel({'l':0, 'm':0}).plot.line(x='Eke', col='Euler');
BLMplot.dXS.real.squeeze().plot.line(x='Eke', col='Euler');
# Plot values normalised by B00 - now set in calculation function
# Plot results with lmPlot(), ordering by Euler sets
# Version with (semi-manual) Euler grouping
daPlot, daPlotpd, legendList, gFig = ep.lmPlot(BLMplot.swap_dims({'Euler':'Labels'}), SFflag = False, eulerGroup = True,
thresType = 'pc', thres = thres,
plotDims = ('Labels','l','m'),
figsize = (15,10))
Differences, L vs. V gauge BLMs
Max difference in BLMs (L-V): 0 (0.5109455268460952+0j)
Plotting data (No filename), pType=a, thres=0.05109455268460952, with Seaborn
[19]:
# Check imaginary components - should be around machine tolerance.
print('Machine tolerance: ', np.finfo(float).eps)
for BLMplot in BLM:
maxImag = BLMplot.imag.max()
print(f'Max imaginary value: {0}', maxImag.data)
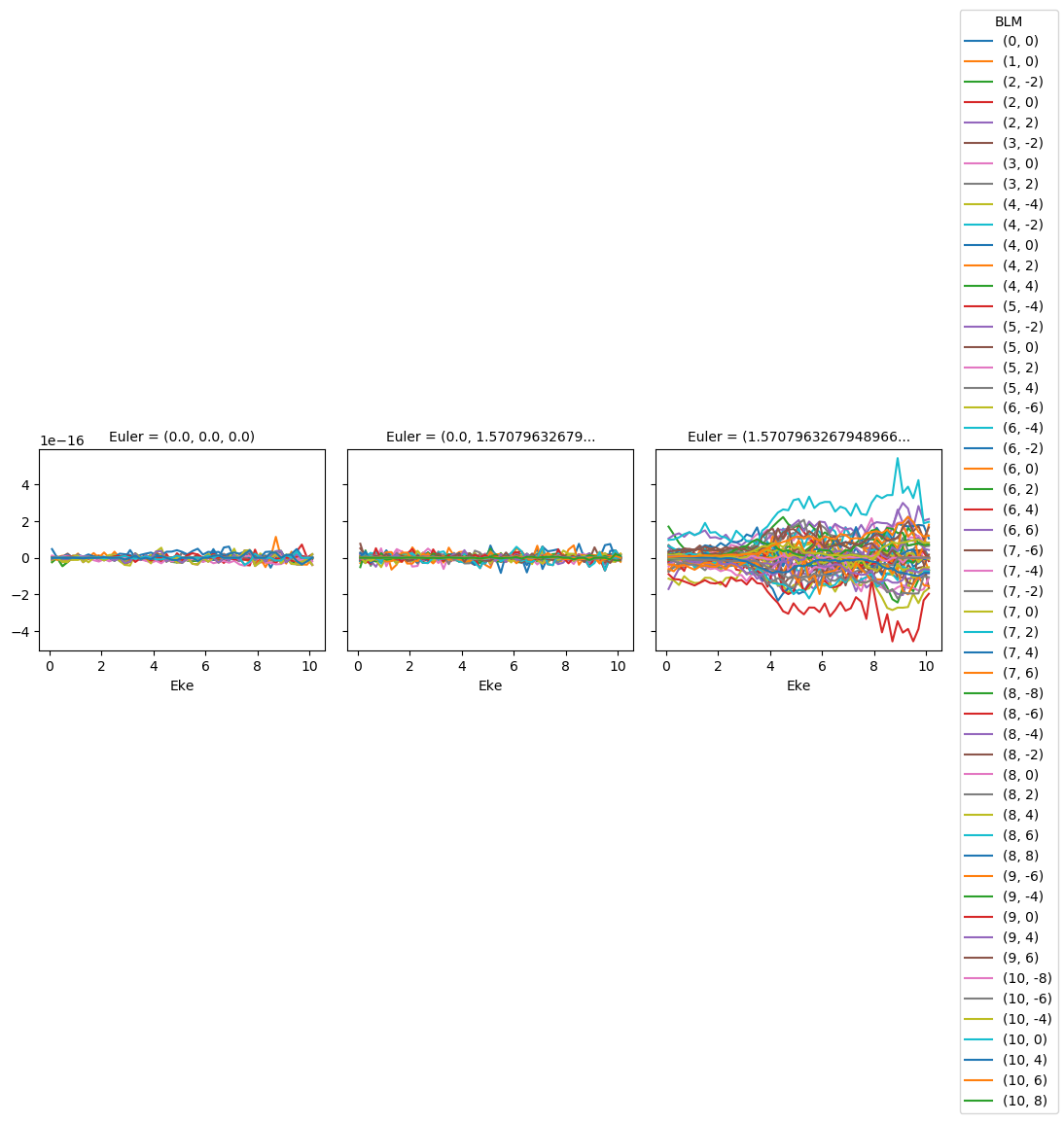
# BLMplot.where(np.abs(BLMplot) > thres, drop = True).imag.squeeze().plot.line(x='Eke', col='Euler');
BLMplot = ep.matEleSelector(BLMplot, thres=thres, dims = 'Eke')
BLMplot.imag.squeeze().plot.line(x='Eke', col='Euler');
Machine tolerance: 2.220446049250313e-16
Max imaginary value: 0 5.429235533647399e-16
Version info¶
Original job details¶
[20]:
print(jobInfo['ePolyScat'][0])
print('Run: ' + jobInfo['Starting'][0].split('at')[1])
ePolyScat Version E3
Run: 2019-03-08 08:32:07.372 (GMT -0500)
ePSproc details¶
[21]:
templateVersion = '0.0.6'
templateDate = '12/01/20'
[22]:
%load_ext version_information
[23]:
%version_information epsproc, xarray
[23]:
| Software | Version |
|---|---|
| Python | 3.7.5 64bit [GCC 7.3.0] |
| IPython | 7.9.0 |
| OS | Linux 5.0.0 36 generic x86_64 with debian buster sid |
| epsproc | 1.2.3 |
| xarray | 0.14.0 |
| Tue Jan 14 14:50:10 2020 EST | |
[24]:
print('Run: {}'.format(timeString.strftime('%Y-%m-%d_%H-%M-%S')))
host = !hostname
print('Host: {}'.format(host[0]))
Run: 2020-01-14_08-50-18
Host: jake
Cite this dataset¶
Hockett, Paul (2019). ePSproc: Aniline Orb 33 (A2) ionization. Dataset on Zenodo. DOI: 10.5281/zenodo.3612509. URL: https://phockett.github.io/ePSdata/aniline/aniline_wf_0.1-10.1eV_orb33_A2.html
Bibtex:
@data{ Aniline Orb 33 (A2) ionization,
title = {ePSproc: Aniline Orb 33 (A2) ionization}
author = {Hockett, Paul},
doi = {10.5281/zenodo.3612509},
publisher = {Zenodo},
year = {2019},
url = {https://phockett.github.io/ePSdata/aniline/aniline_wf_0.1-10.1eV_orb33_A2.html}
}
See citation notes on ePSdata for further details.
