This page was auto-generated from a Jupyter notebook: about.
Problems? Please raise any issues on Github.
About ePSdata¶
21/01/20
Overview¶
ePolyScat (ePS) is an open-source tool for numerical computation of electron-molecule scattering & photoionization by Lucchese & coworkers. For more details try:
Calculation of low-energy elastic cross sections for electron-CF4 scattering, F. A. Gianturco, R. R. Lucchese, and N. Sanna, J. Chem. Phys. 100, 6464 (1994), http://dx.doi.org/10.1063/1.467237
Cross section and asymmetry parameter calculation for sulfur 1s photoionization of SF6, A. P. P. Natalense and R. R. Lucchese, J. Chem. Phys. 111, 5344 (1999), http://dx.doi.org/10.1063/1.479794
ePSproc is an open-source tool for post-processing & visualisation of ePS results, aimed primarily at photoionization studies.
Ongoing documentation is on Read the Docs.
Source code is available on Github.
For more background, see the software metapaper for the original release of ePSproc (Aug. 2016): ePSproc: Post-processing suite for ePolyScat electron-molecule scattering calculations, on Authorea or arXiv 1611.04043.
ePSdata is an open-data/open-science collection of ePS + ePSproc results.
ePSdata collects ePS datasets, post-processed via ePSproc (Python) in Jupyter notebooks, for a full open-data/open-science transparent pipeline.
ePSdata is currently (Jan 2020) collecting existing calculations from 2010 - 2019, from the femtolabs at NRC, with one notebook per ePS job.
In future, ePSdata pages will be automatically generated from ePS jobs (via the ePSman toolset, currently in development), for immediate dissemination to the research community.
Source notebooks are available on the Github project pages, and notebooks + datasets via Zenodo repositories (one per dataset). Each notebook + dataset is given a Zenodo DOI for full traceability, and notebooks are versioned on Github.
Note: ePSdata may also be linked or mirrored on the existing ePolyScat Collected Results OSF project, but will effectively supercede those pages.
All results are released under Creative Commons Attribution-NonCommercial-ShareAlike 4.0 (CC BY-NC-SA 4.0) license, and are part of our ongoing Open Science initiative.


Demos¶
For an introduction to ePS and ePSproc, see:
Sample ePS jobs from the ePolyScat website and manual
Demos for ePSproc:
:math:beta_{lm}` calculations demo <https://epsproc.readthedocs.io/en/latest/ePSproc_BLM_calc_demo_Sept2019_rst/ePSproc_BLM_calc_demo_Sept2019.html>`__
(More demos coming soon.)
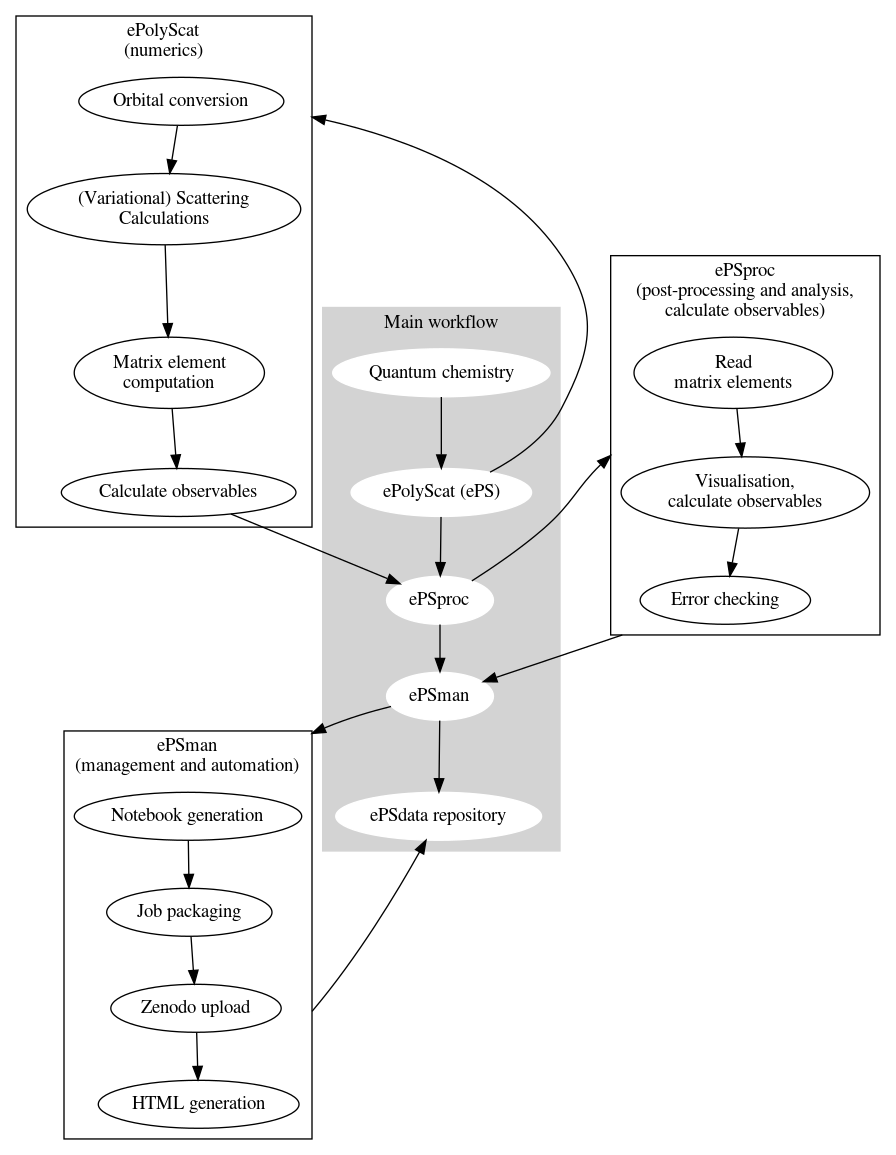
Workflow¶
The general workflow for photoionization calculations plus post-processing is shown below. This pipeline involves a range of code suites, as shown in the main workflow; some additional details are also illustrated.
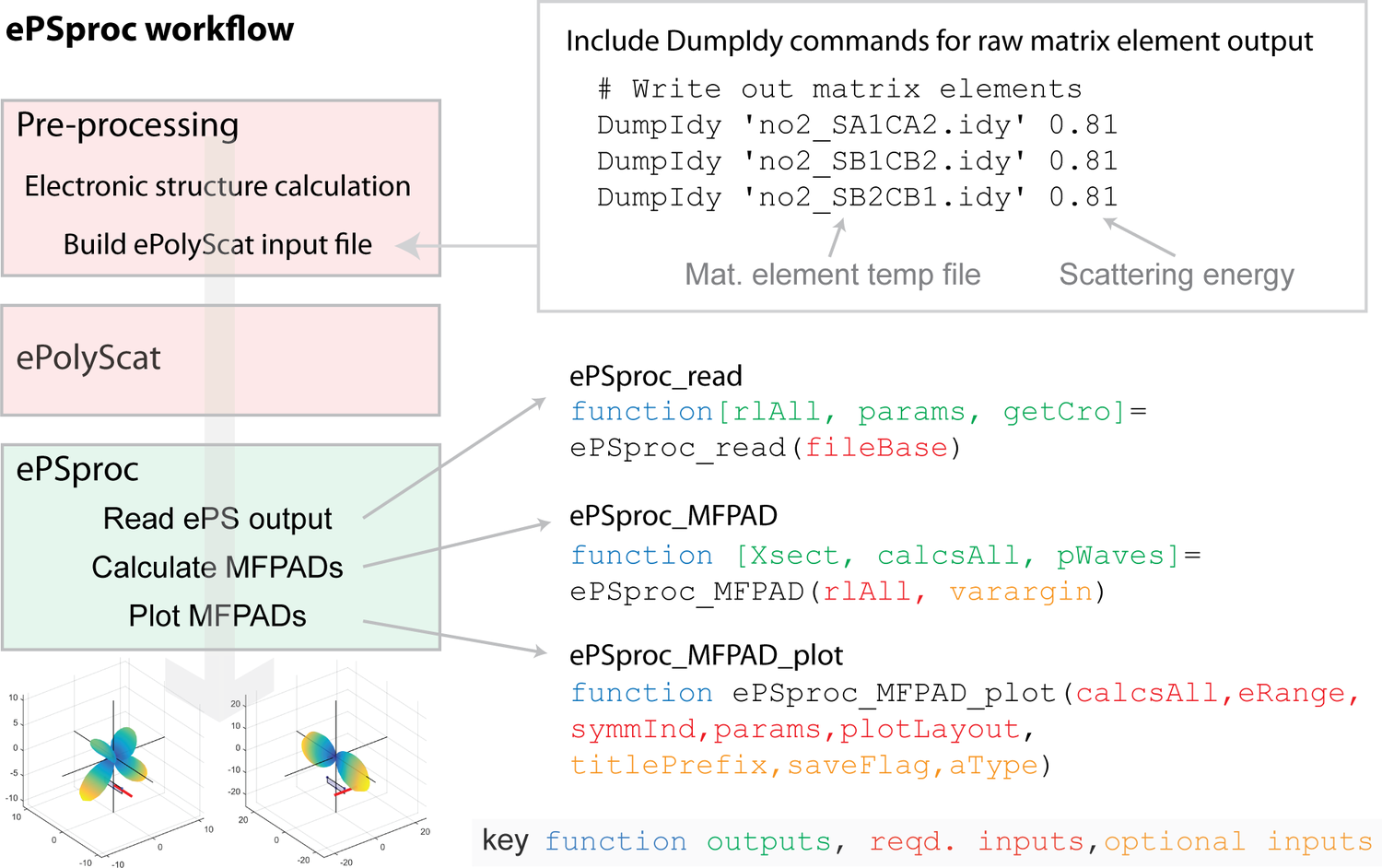
The basic ePSproc workflow (from the original software metapaper is shown below: essentially, the ePS output file is parsed for photoionization matrix elements, and ancillary data; the matrix elements are the used to calculate properties of interest, such as photoionziation cross-sections and MF-PADs. For ePSdata, this workflow is applied to each ePS dataset, with a Jupyter notebook as a template. The completed analysis & dataset is then uploaded to Zenodo, and HTML version to ePSdata.
Citation & acknowledgements¶
Each notebook & dataset has a DOI, see the citations page for further details.
Motivation¶
Scientific¶
Platform development for quantum metrology with photoelectrons; see, for example:
Theory and method development.
Bridge-building and method unification between theory and experiment.
Lowering the barrier to entry for photoionization studies.
Sharing and disemination of computational results and data, which would not otherwise be subject to “traditional” publication, and just sit on a hard-drive.
General good scientific practice and open science…
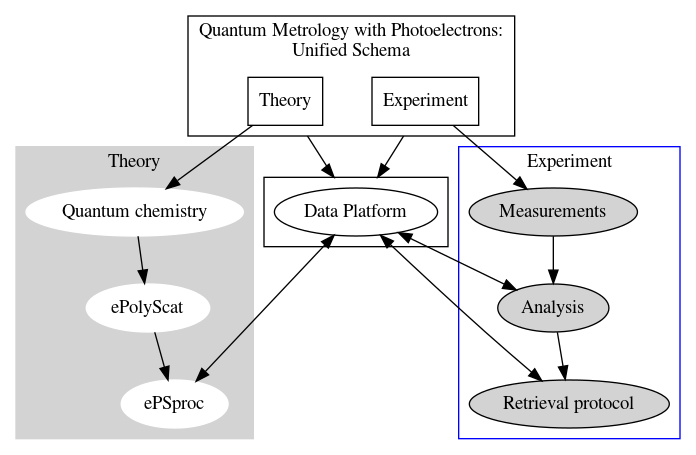
Platform development for quantum metrology with photoelectrons: figures below show existing and new platform schematics. In the former case, theory and experiment are treated separately, and compared at the level of observables; in the latter lower-level comparisons are possible and aid analysis. 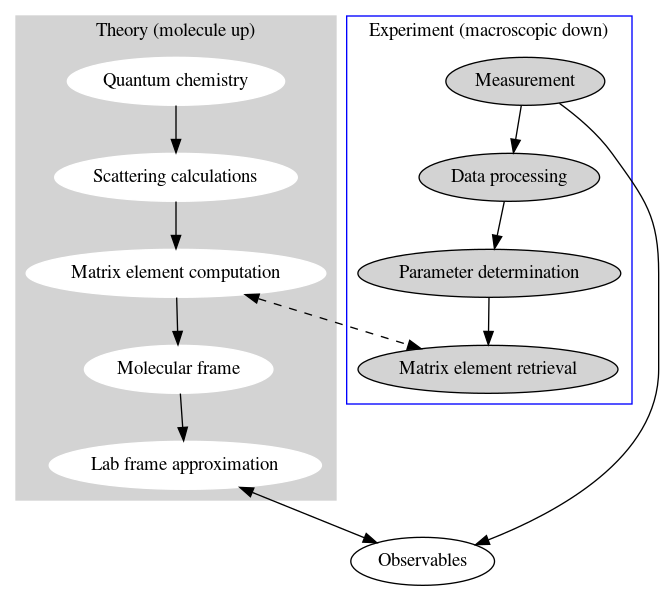
Science policy/open science¶
All things open: open data, open science, open source software. (See further reading below, and also the Open Science Initiative).
Reproducibility, and avoiding the reproducibility crises.
Transparency, and making full computational results stacks available.
Building a community resource.
Some further reading on these topics, in no particular order:
Promoting an open research culture. Nosek et. al. (2015). Science, 348(6242), 1422–1425.
Publish your computer code: it is good enough. Barnes, N. (2010). Nature, 467(7317), 753.
For more general discussion, tools and further resources, see, for example:
Reproducible Research and Data Analysis module at Open Science MOOC (includes many more tools, links and references, many other modules are available too).
Notes¶
Web pages are auto-generated from source Jupyter notebooks: please raise any issues on Github.
Figures may appear small due to HTML template constraints, but opening in a new window or downloading will provide the high-res source. (This will be fixed at some point in the future…)