4.1. Excited state wavepacket: density matrices and spatial distributions (legacy version)#
From the simulated electronic wavepacket, we can analyse the properties in density matrix and real-space representations.
Basic “Legacy version” uses basic code from Alignment 3 notebook. (This page.)
Update version, uses code from this project and ePSproc for analysis. See Excited state wavepacket: density matrices and spatial distributions (updated version).
From prior work and data:
Forbes, R. et al. (2018) ‘Quantum-beat photoelectron-imaging spectroscopy of Xe in the VUV’, Physical Review A, 97(6), p. 063417. Available at: https://doi.org/10.1103/PhysRevA.97.063417. arXiv: http://arxiv.org/abs/1803.01081, Authorea (original HTML version): https://doi.org/10.22541/au.156045380.07795038
Data (OSF): https://osf.io/ds8mk/
Quantum Metrology with Photoelectrons (Github repo), particularly the Alignment 3 notebook. Functions from this notebook have been incorporated in the current project, under
qbanalysis.hyperfine.
4.1.1. Imports#
# For testing
# %load_ext autoreload
# %autoreload 2
# Load packages
# Main functions used herein from qbanalysis.hyperfine
from qbanalysis.hyperfine import *
import numpy as np
from epsproc.sphCalc import setBLMs
from pathlib import Path
dataPath = Path('/tmp/xe_analysis')
# dataTypes = ['BLMall', 'BLMerr', 'BLMerrCycle'] # Read these types, should just do dir scan here.
# # Read from HDF5/NetCDF files
# # TO FIX: this should be identical to loadFinalDataset(dataPath), but gives slightly different plots - possibly complex/real/abs confusion?
# dataDict = {}
# for item in dataTypes:
# dataDict[item] = IO.readXarray(fileName=f'Xe_dataset_{item}.nc', filePath=dataPath.as_posix()).real
# dataDict[item].name = item
# Read from raw data files
from qbanalysis.dataset import loadFinalDataset
dataDict = loadFinalDataset(dataPath)
# Use Pandas and load Xe local data (ODS)
# These values were detemermined from the experimental data as detailed in ref. [4].
# from qbanalysis.dataset import loadXeProps
# xeProps = loadXeProps()
# Load adv. fit data
from qbanalysis.dataset import loadAdvFit
xePropsFit, xeParamsFit, paramsUDict = loadAdvFit()
2025-06-03 16:52:26.838 | INFO | qbanalysis.config:<module>:11 - PROJ_ROOT path is: /home/runner/work/Quantum-Beat_Photoelectron-Imaging_Spectroscopy_of_Xe_in_the_VUV/Quantum-Beat_Photoelectron-Imaging_Spectroscopy_of_Xe_in_the_VUV
* sparse not found, sparse matrix forms not available.
* natsort not found, some sorting functions not available.
* Setting plotter defaults with epsproc.basicPlotters.setPlotters(). Run directly to modify, or change options in local env.
* Set Holoviews with bokeh.
* pyevtk not found, VTK export not available.
* pyshtools not found, SHtools functions not available. If required, run "pip install pyshtools" or "conda install -c conda-forge pyshtools" to install.
2025-06-03 16:52:47.648 | INFO | qbanalysis.hyperfine:<module>:31 - Using uncertainties modules, Sympy maths functions will be forced to float outputs.
2025-06-03 16:52:47.723 | INFO | qbanalysis.dataset:loadDataset:268 - Loaded data cpBasex_results_cycleSummed_rot90_quad1_ROI_results_with_FT_NFFT1024_hanningWindow_270717.mat.
2025-06-03 16:52:47.772 | INFO | qbanalysis.dataset:loadDataset:268 - Loaded data cpBasex_results_allCycles_ROIs_with_FTs_NFFT1024_hanningWindow_270717.mat.
/opt/hostedtoolcache/Python/3.10.11/x64/lib/python3.10/site-packages/xarray/core/concat.py:500: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
common_dims = tuple(pd.unique([d for v in vars for d in v.dims]))
2025-06-03 16:52:48.104 | INFO | qbanalysis.dataset:loadFinalDataset:244 - Processed data to Xarray OK.
2025-06-03 16:52:48.154 | INFO | qbanalysis.dataset:loadAdvFit:123 - Loaded Xe adv. fit data from /home/runner/work/Quantum-Beat_Photoelectron-Imaging_Spectroscopy_of_Xe_in_the_VUV/Quantum-Beat_Photoelectron-Imaging_Spectroscopy_of_Xe_in_the_VUV/dataLocal/xeAdvFit.h5.
# v2 pkg
from qbanalysis.adv_fitting import *
2025-06-03 16:52:48.172 | INFO | qbanalysis.basic_fitting:<module>:21 - Using uncertainties modules, Sympy maths functions will be forced to float outputs.
2025-06-03 16:52:48.172 | INFO | qbanalysis.adv_fitting:<module>:29 - Using uncertainties modules, Sympy maths functions will be forced to float outputs.
Show code cell content
# # Hide future warnings from Xarray concat for fitting on some platforms
# import warnings
# # warnings.filterwarnings('ignore') # ALL WARNINGS
# # warnings.filterwarnings('ignore', category=DeprecationWarning)
# warnings.filterwarnings('ignore', category=FutureWarning)
4.1.2. Rerun model from loaded parameters#
Generate data and verify.
# Recalc model with uncertainties & plot...
# NOTE: currently doesn't include uncertainties on t-coord.
# TODO: add labels and fix ledgend in layout
from qbanalysis.plots import plotFinalDatasetBLMt
plotOpts = {'width':800}
calcDict = calcAdvFitModel(paramsUDict, xePropsFit=xePropsFit, dataDict=dataDict)
# plotHyperfineModel(calcDict['ionization'],overlay=['ROI']).layout('l')
# To fix layout issues, treat l separately...
l2 = (plotFinalDatasetBLMt(**dataDict) * plotHyperfineModel(calcDict['ionization'],overlay=['ROI'])).select(l=2)
l4 = (plotFinalDatasetBLMt(**dataDict) * plotHyperfineModel(calcDict['ionization'],overlay=['ROI'])).select(l=4)
(l2.overlay('l').opts(title="l2", **plotOpts) + l4.overlay('l').opts(title="l4", **plotOpts)).cols(1)
/opt/hostedtoolcache/Python/3.10.11/x64/lib/python3.10/site-packages/xarray/core/concat.py:500: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
common_dims = tuple(pd.unique([d for v in vars for d in v.dims]))
/opt/hostedtoolcache/Python/3.10.11/x64/lib/python3.10/site-packages/xarray/core/concat.py:500: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
common_dims = tuple(pd.unique([d for v in vars for d in v.dims]))
/opt/hostedtoolcache/Python/3.10.11/x64/lib/python3.10/site-packages/xarray/core/concat.py:500: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
common_dims = tuple(pd.unique([d for v in vars for d in v.dims]))
/opt/hostedtoolcache/Python/3.10.11/x64/lib/python3.10/site-packages/xarray/core/concat.py:500: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
common_dims = tuple(pd.unique([d for v in vars for d in v.dims]))
/opt/hostedtoolcache/Python/3.10.11/x64/lib/python3.10/site-packages/xarray/core/concat.py:500: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
common_dims = tuple(pd.unique([d for v in vars for d in v.dims]))
/opt/hostedtoolcache/Python/3.10.11/x64/lib/python3.10/site-packages/xarray/core/concat.py:500: FutureWarning: unique with argument that is not not a Series, Index, ExtensionArray, or np.ndarray is deprecated and will raise in a future version.
common_dims = tuple(pd.unique([d for v in vars for d in v.dims]))
# calcDict['modelDict']['129Xe'] #.keys()
4.1.3. Density matrix representations#
Use old-code for basic outputs.
See updated ePSproc/PEMtk density matrix codes for updated plots.
calcDict['modelDict']['129Xe'].t #attrs['states'] #.TKQ[0].data.item()[0]
<xarray.DataArray 't' (t: 97)>
array([-70., -60., -50., -40., -30., -20., -10., 0., 10., 20., 30., 40.,
50., 60., 70., 80., 90., 100., 110., 120., 130., 140., 150., 160.,
170., 180., 190., 200., 210., 220., 230., 240., 250., 260., 270., 280.,
290., 300., 310., 320., 330., 340., 350., 360., 370., 380., 390., 400.,
410., 420., 430., 440., 450., 460., 470., 480., 490., 500., 510., 520.,
530., 540., 550., 560., 570., 580., 590., 600., 610., 620., 630., 640.,
650., 660., 670., 680., 690., 700., 710., 720., 730., 740., 750., 760.,
770., 780., 790., 800., 810., 820., 830., 840., 850., 860., 870., 880.,
890.])
Coordinates:
* t (t) float64 -70.0 -60.0 -50.0 -40.0 ... 860.0 870.0 880.0 890.0
Attributes:
units: psdef pJpNpJNXR(Jp,J,TKQXR):
"""
Compute pJpNpJN(Jp,J,TKQ)
\begin{equation}
\langle J'N'|\hat{\rho}|JN\rangle=\sum_{N'N}(-1)^{J'-N'}(2K+1)^{1/2}\left(\begin{array}{ccc}
J' & J & K\\
N' & -N & -Q
\end{array}\right)\left\langle T(J',J)_{KQ}^{\dagger}\right\rangle
\end{equation}
# Define density matrix p(Jp,Np,J,N) from TKQ - general version, eqn. 4.34 in Blum (p125)
# Uses TKQ tensor values (list)
"""
# Set data for legacy code
KQ = TKQXR.TKQ
TKQ = TKQXR.data
if unFlag:
TKQ = unumpy.nominal_values(TKQ)
Jmax = max(J,Jp)
Pmm = np.zeros((2*Jmax+1,2*Jmax+1))
for Mp in range(-Jp,Jp+1):
for M in range(-J,J+1):
for row in range(KQ.shape[0]):
K = KQ[row].data.item()[0]
Q = KQ[row].data.item()[1]
Pmm[Mp+Jp][M+J] += (-1)**(Jp-Mp)*sqrt(2*K+1)*wigner_3j(Jp,J,K,Mp,-M,-Q)*TKQ[row]
return Pmm
# Basic case - plot single t-point, per Alignment-1 notebook.
# Set data
TKQ = calcDict['modelDict']['129Xe'][10]
# Set case
# Jp = 2 # Set angular momenta - assume a singe J-state, hence Jp=J
# J = Jp
isoKey = '129Xe'
JFlist = calcDict['modelDict'][isoKey].attrs['states']['JFlist']
Jf = np.int(JFlist[0][0]) # Final state J
# Set terms for legacy code
J = Jf
Jp = Jf
pmm = pJpNpJNXR(Jf,Jf,TKQ) # Determine pmm
pmm
/tmp/ipykernel_2590/1516625454.py:11: DeprecationWarning: `np.int` is a deprecated alias for the builtin `int`. To silence this warning, use `int` by itself. Doing this will not modify any behavior and is safe. When replacing `np.int`, you may wish to use e.g. `np.int64` or `np.int32` to specify the precision. If you wish to review your current use, check the release note link for additional information.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
Jf = np.int(JFlist[0][0]) # Final state J
array([[0.07957993, 0. , 0. ],
[0. , 0.17417348, 0. ],
[0. , 0. , 0.07957993]])
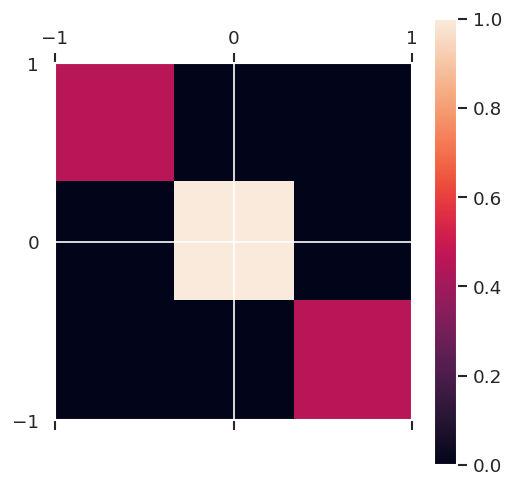
print('Original ensemble')
print('Trace(pmm) = {:f}'.format(np.trace(pmm)))
# print(TKQ)
plt.matshow(pmm/np.amax(pmm), extent = (-Jp,Jp,-J,J), aspect = 'equal')
plt.colorbar()
plt.show()
Original ensemble
Trace(pmm) = 0.333333
calcDict.keys()
dict_keys(['xData', 'xePropsFit', 'dataDict', 'trange', 'fitFlag', 'returnType', 'modelDict', 'modelDictSum', 'modelDA', 'modelSum', 'dataIn', 'modelIn', 'res', 'iso', 'dataCol', 'decay', 'ionization'])
Quick plots of \(\rho(t)\)
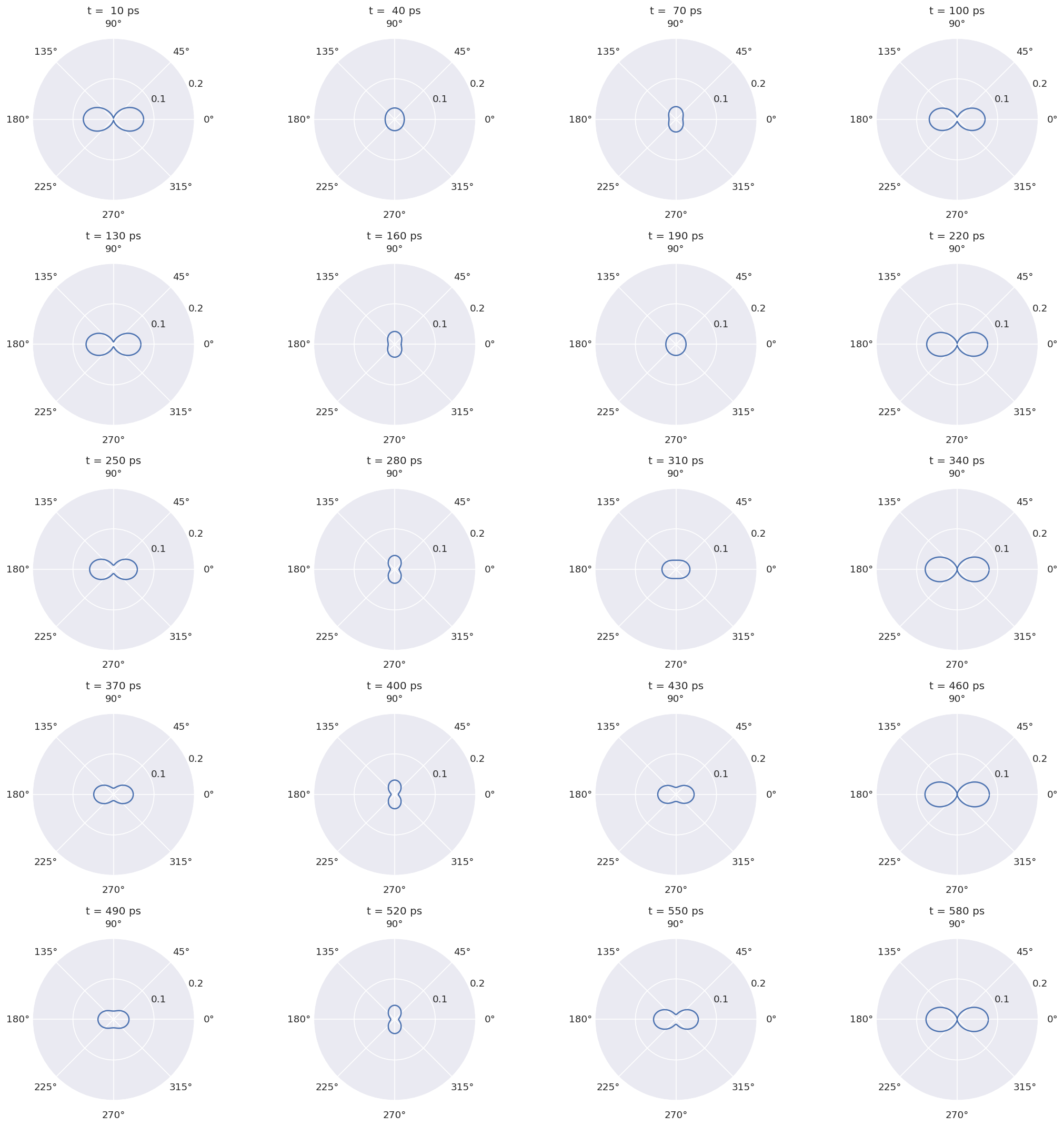
Note period ~110ps for 129Xe, ~600ps for 131Xe.
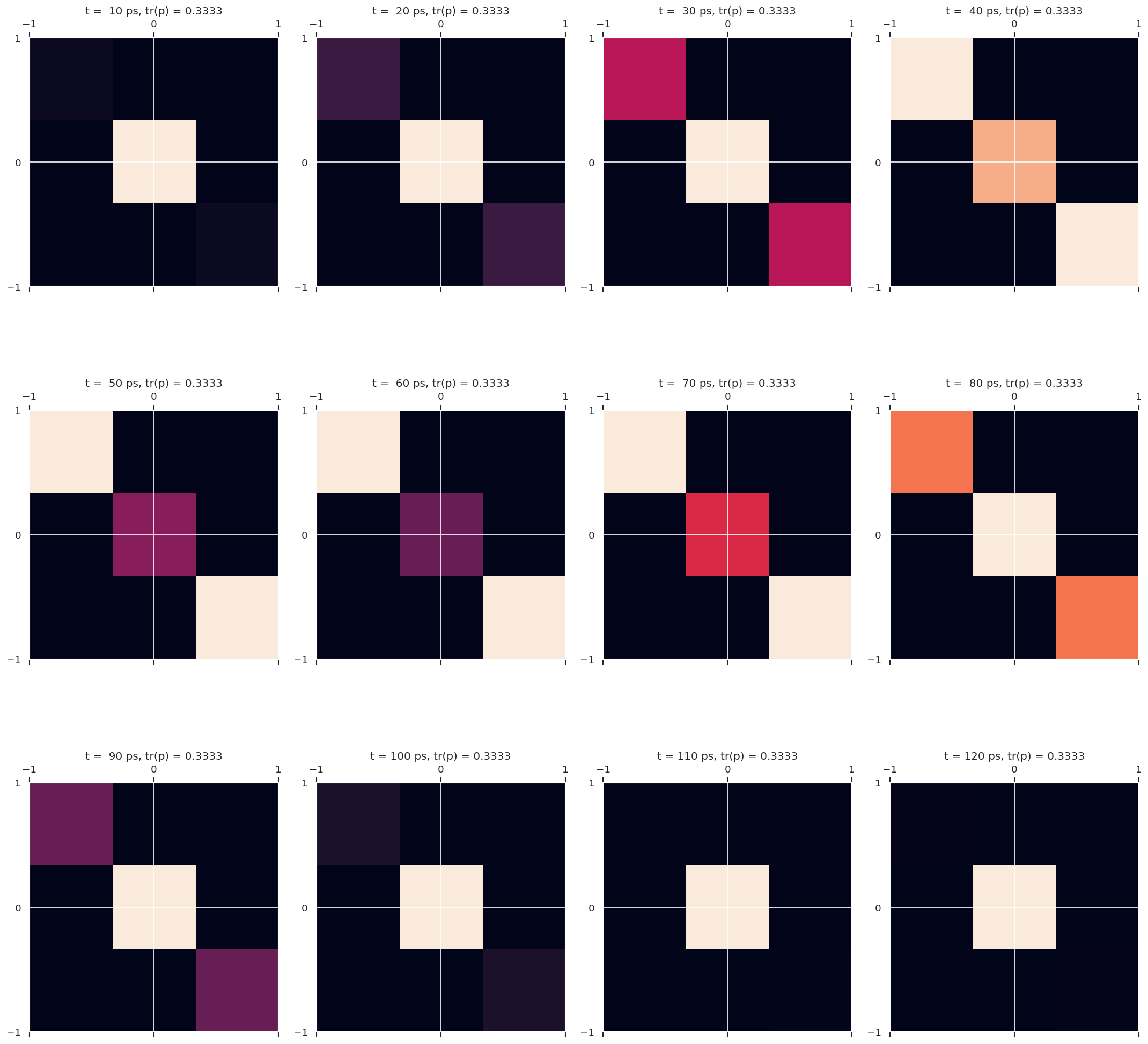
4.1.3.1. Density matrices for \(^{129}Xe\)#
# Plot pmm(t) using modified version of old code...
tStart = 8
tEnd = 20
# tEnd = round(tIn.shape[0]/4)
tStep = 1
# Set up subplots. For polar use, see example at https://matplotlib.org/gallery/subplots_axes_and_figures/subplots_demo.html
sPlots = [3,4]
fig, axes = plt.subplots(sPlots[0], sPlots[1], figsize=(18, 18))
n = m = 0
# Set final state parameters by isotope
# JFlist = JF131
# Jf = np.int(JFlist[0][0]) # Final state J
isoKey = '129Xe'
JFlist = calcDict['modelDict'][isoKey].attrs['states']['JFlist']
# Jf = np.int(JFlist[0][0]) # Final state J
Jf = calcDict['modelDict'][isoKey].attrs['states']['Jf']
Ji = calcDict['modelDict'][isoKey].attrs['states']['Ji']
tIn = calcDict['modelDict'][isoKey].t.data
print('p(Jf;t) for (Ji,Jf) = ({0},{1})'.format(Ji,Jf))
for tPlot in range(tStart,tEnd,tStep):
# Calculate
# TKQin = np.vstack((TKQ[:,0:2].T,TJt[:,tPlot])).T
# pmm = pJpNpJN(Jf,Jf,TKQin) # Determine pmm
# print(f"tPlot={tPlot}, t={tIn[tPlot]}")
# Set data
TKQin = calcDict['modelDict'][isoKey][tPlot]
pmm = pJpNpJNXR(Jf,Jf,TKQin) # Determine pmm
# Singe polar plot
# plt.polar(np.concatenate((tList, tList+pi)),np.concatenate((Ytp, Ytp)),fig=fig, ax=axes[n, m]) # Manual fix to symmetry for theta = 0:2pi
# Polar subplot, with bounds checking
if (m+1)*(n+1)>(sPlots[0]*sPlots[1]):
pass
elif (n+1)>sPlots[0]:
pass
else:
axes[n,m].matshow(pmm/np.trace(pmm), extent = (-Jf,Jf,-Jf,Jf), aspect = 'equal')
# axes[n,m].set_title('t = {:3.0f} ps, tr(p) = {:1.4f}'.format(tIn[tPlot]/1e-12,np.trace(pmm)))
axes[n,m].set_title('t = {:3.0f} ps, tr(p) = {:1.4f}'.format(tIn[tPlot],np.trace(pmm)))
# Subplot indexing
m += 1
if m >= sPlots[1]:
m = 0
n += 1
# plt.subplots_adjust(hspace=0.5) # Fix overlapping titles
fig.tight_layout() # Or use tight_layout.
plt.show()
p(Jf;t) for (Ji,Jf) = (0,1)
4.1.3.2. Density matrices for \(^{131}Xe\)#
# Plot pmm(t) using modified version of old code...
tStart = 8
tEnd = 90
# tEnd = round(tIn.shape[0]/4)
tStep = 4
# Set up subplots. For polar use, see example at https://matplotlib.org/gallery/subplots_axes_and_figures/subplots_demo.html
sPlots = [4,4]
fig, axes = plt.subplots(sPlots[0], sPlots[1], figsize=(18, 18))
n = m = 0
# Set final state parameters by isotope
# JFlist = JF131
# Jf = np.int(JFlist[0][0]) # Final state J
isoKey = '131Xe'
JFlist = calcDict['modelDict'][isoKey].attrs['states']['JFlist']
# Jf = np.int(JFlist[0][0]) # Final state J
Jf = calcDict['modelDict'][isoKey].attrs['states']['Jf']
Ji = calcDict['modelDict'][isoKey].attrs['states']['Ji']
tIn = calcDict['modelDict'][isoKey].t.data
print('p(Jf;t) for (Ji,Jf) = ({0},{1})'.format(Ji,Jf))
for tPlot in range(tStart,tEnd,tStep):
# Calculate
# TKQin = np.vstack((TKQ[:,0:2].T,TJt[:,tPlot])).T
# pmm = pJpNpJN(Jf,Jf,TKQin) # Determine pmm
# print(f"tPlot={tPlot}, t={tIn[tPlot]}, [n,m]={n,m}")
# Set data
TKQin = calcDict['modelDict'][isoKey][tPlot]
pmm = pJpNpJNXR(Jf,Jf,TKQin) # Determine pmm
# Singe polar plot
# plt.polar(np.concatenate((tList, tList+pi)),np.concatenate((Ytp, Ytp)),fig=fig, ax=axes[n, m]) # Manual fix to symmetry for theta = 0:2pi
# Polar subplot, with bounds checking
# if (m+1)+((n-1)*sPlots[0])>(sPlots[0]*sPlots[1]):
if (m+1)*(n+1)>(sPlots[0]*sPlots[1]):
pass
elif (n+1)>sPlots[0]:
pass
else:
axes[n,m].matshow(pmm/np.trace(pmm), extent = (-Jf,Jf,-Jf,Jf), aspect = 'equal')
# axes[m,n].matshow(pmm/np.trace(pmm), extent = (-Jf,Jf,-Jf,Jf), aspect = 'equal')
# axes[n,m].set_title('t = {:3.0f} ps, tr(p) = {:1.4f}'.format(tIn[tPlot]/1e-12,np.trace(pmm)))
axes[n,m].set_title('t = {:3.0f} ps, tr(p) = {:1.4f}'.format(tIn[tPlot],np.trace(pmm)))
# axes[m,n].set_title('t = {:3.0f} ps, tr(p) = {:1.4f}'.format(tIn[tPlot],np.trace(pmm)))
# Subplot indexing
m += 1
if m >= sPlots[1]:
m = 0
n += 1
# plt.subplots_adjust(hspace=0.5) # Fix overlapping titles
fig.tight_layout() # Or use tight_layout.
plt.show()
p(Jf;t) for (Ji,Jf) = (0,1)

4.1.4. Electronic state distribution/alignment#
Expand \(T_{K,Q}\) in spherical harmonics for real-space electronic state density.
# Function to sum Ylm from a list, with optional normalisation.
# Include additional 3j term to implement eqn. 101, for real-space W(theta,phi) representation.
def sphSumTKQXR(AXR, J, norm = 1.0):
Atp = 0
thres = 1E-5
# Set data for legacy code
AKQ = AXR.TKQ
A = AXR.data
if unFlag:
A = unumpy.nominal_values(A)
# Loop over rows in input & add YKQ terms (should be able to convert to list comprehension for brevity)
for row in range(AKQ.shape[0]):
if np.absolute(A[row]) > thres:
K = AKQ[row].data.item()[0]
Q = AKQ[row].data.item()[1]
angMomTerm = (-1)**J * (2*J+1) * wigner_3j(J,J,K,0,0,0)
Atp += angMomTerm*Ynm(np.int(K),np.int(Q),theta,phi) * A[row]/norm # Add TKQ*Y(K,Q) term
return Atp*sqrt(1/(4*pi))
# Jmax = max(J,Jp)
# Pmm = np.zeros((2*Jmax+1,2*Jmax+1))
# for Mp in range(-Jp,Jp+1):
# for M in range(-J,J+1):
# for row in range(KQ.shape[0]):
# K = KQ[row].data.item()[0]
# Q = KQ[row].data.item()[1]
# Pmm[Mp+Jp][M+J] += (-1)**(Jp-Mp)*sqrt(2*K+1)*wigner_3j(Jp,J,K,Mp,-M,-Q)*TKQ[row]
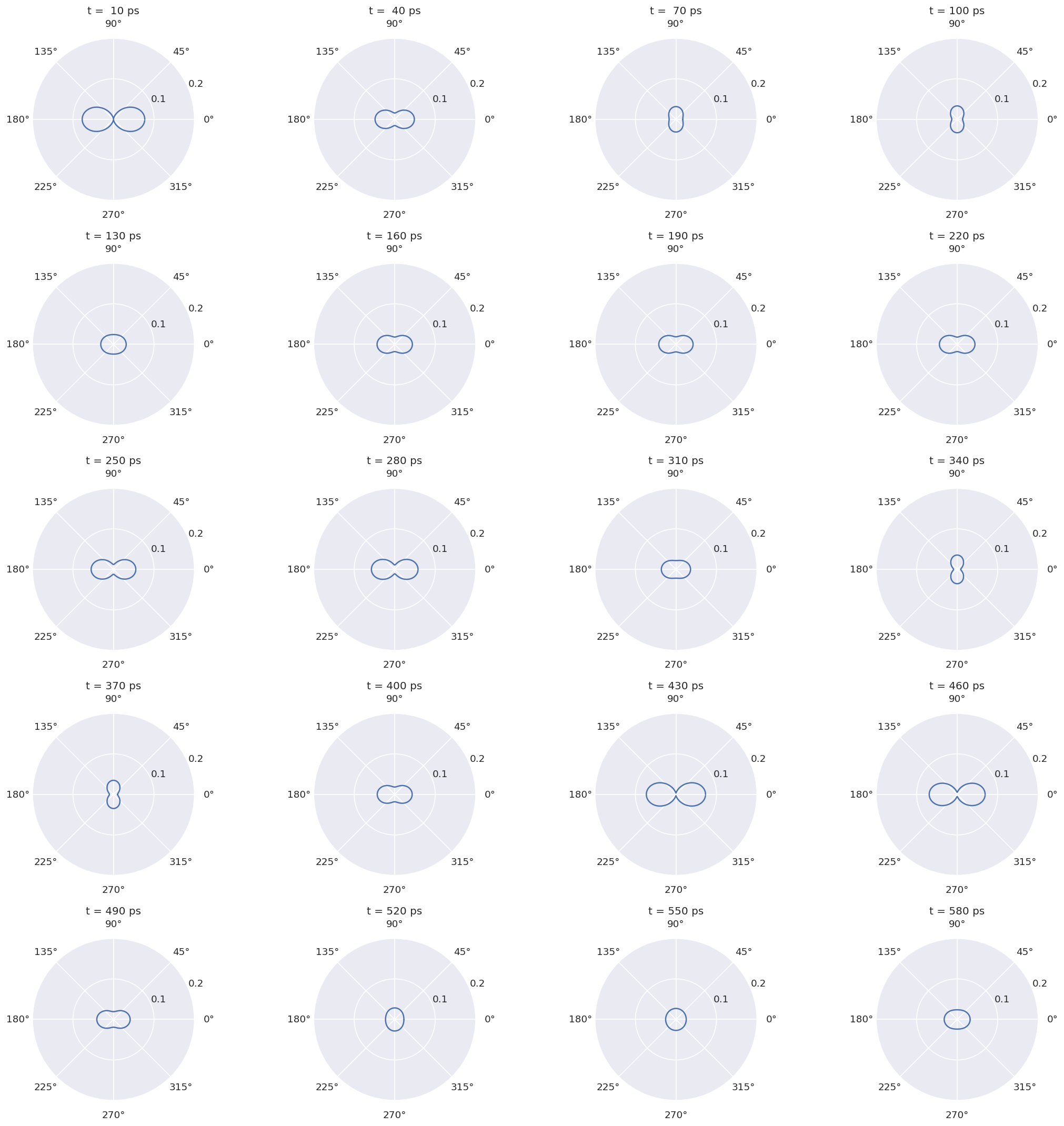
4.1.4.1. Spatial distributions for \(^{129}Xe\)#
# Define temporal points to plot as index into tIn
tStart = 8
tEnd = 66
# tEnd = round(tIn.shape[0]/4)
tStep = 3
# Define theta values for plot
tList = np.arange(0,pi,0.05)
# print('T(J;t) for (Ji,Jf) = ({0},{1}), p = {2}'.format(Ji,Jf,p))
# print('\n At 1-photon abs.')
# print(TKQ)
# Set final state parameters by isotope
# JFlist = JF131
# Jf = np.int(JFlist[0][0]) # Final state J
isoKey = '129Xe'
JFlist = calcDict['modelDict'][isoKey].attrs['states']['JFlist']
# Jf = np.int(JFlist[0][0]) # Final state J
Jf = calcDict['modelDict'][isoKey].attrs['states']['Jf']
Ji = calcDict['modelDict'][isoKey].attrs['states']['Ji']
tIn = calcDict['modelDict'][isoKey].t.data
print('\n At various t...')
# Set up subplots. For polar use, see example at https://matplotlib.org/gallery/subplots_axes_and_figures/subplots_demo.html
sPlots = [5,4]
fig, axes = plt.subplots(sPlots[0], sPlots[1], figsize=(18, 18), subplot_kw=dict(projection='polar'))
# fig.tight_layout()
n = m = 0
for tPlot in range(tStart,tEnd,tStep):
# Set data
TKQin = calcDict['modelDict'][isoKey][tPlot]
# pmm = pJpNpJNXR(Jf,Jf,TKQin) # Determine pmm
# Calculate
# TKQin = np.vstack((TKQ[:,0:2].T,TJt[:,tPlot])).T
Atp = sphSumTKQXR(TKQin, Jf,) # norm = TKQin)
Ytp = sphNList(Atp,tList)
# Singe polar plot
# plt.polar(np.concatenate((tList, tList+pi)),np.concatenate((Ytp, Ytp)),fig=fig, ax=axes[n, m]) # Manual fix to symmetry for theta = 0:2pi
# Polar subplot, with bounds checking
# Polar subplot, with bounds checking
# if (m+1)+((n-1)*sPlots[0])>(sPlots[0]*sPlots[1]):
if (m+1)*(n+1)>(sPlots[0]*sPlots[1]):
pass
elif (n+1)>sPlots[0]:
pass
else:
axes[n,m].plot(np.concatenate((tList, tList+pi)),np.concatenate((Ytp, Ytp)))
axes[n,m].set_title('t = {:3.0f} ps'.format(tIn[tPlot]))
axes[n,m].set_rticks([0.1, 0.2]) # Reduce radial ticks
# Subplot indexing
m += 1
if m >= sPlots[1]:
m = 0
n += 1
# plt.subplots_adjust(hspace=0.5) # Fix overlapping titles
fig.tight_layout() # Or use tight_layout.
plt.show()
At various t...
/tmp/ipykernel_2590/3300663484.py:20: DeprecationWarning: `np.int` is a deprecated alias for the builtin `int`. To silence this warning, use `int` by itself. Doing this will not modify any behavior and is safe. When replacing `np.int`, you may wish to use e.g. `np.int64` or `np.int32` to specify the precision. If you wish to review your current use, check the release note link for additional information.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
Atp += angMomTerm*Ynm(np.int(K),np.int(Q),theta,phi) * A[row]/norm # Add TKQ*Y(K,Q) term
4.1.4.2. Spatial distributions for \(^{131}Xe\)#
# Define temporal points to plot as index into tIn
tStart = 8
tEnd = 66
# tEnd = round(tIn.shape[0]/4)
tStep = 3
# Define theta values for plot
tList = np.arange(0,pi,0.05)
# print('T(J;t) for (Ji,Jf) = ({0},{1}), p = {2}'.format(Ji,Jf,p))
# print('\n At 1-photon abs.')
# print(TKQ)
# Set final state parameters by isotope
# JFlist = JF131
# Jf = np.int(JFlist[0][0]) # Final state J
isoKey = '131Xe'
JFlist = calcDict['modelDict'][isoKey].attrs['states']['JFlist']
# Jf = np.int(JFlist[0][0]) # Final state J
Jf = calcDict['modelDict'][isoKey].attrs['states']['Jf']
Ji = calcDict['modelDict'][isoKey].attrs['states']['Ji']
tIn = calcDict['modelDict'][isoKey].t.data
print('\n At various t...')
# Set up subplots. For polar use, see example at https://matplotlib.org/gallery/subplots_axes_and_figures/subplots_demo.html
sPlots = [5,4]
fig, axes = plt.subplots(sPlots[0], sPlots[1], figsize=(18, 18), subplot_kw=dict(projection='polar'))
n = m = 0
for tPlot in range(tStart,tEnd,tStep):
# Set data
TKQin = calcDict['modelDict'][isoKey][tPlot]
# pmm = pJpNpJNXR(Jf,Jf,TKQin) # Determine pmm
# Calculate
# TKQin = np.vstack((TKQ[:,0:2].T,TJt[:,tPlot])).T
Atp = sphSumTKQXR(TKQin, Jf,) # norm = TKQin)
Ytp = sphNList(Atp,tList)
# Singe polar plot
# plt.polar(np.concatenate((tList, tList+pi)),np.concatenate((Ytp, Ytp)),fig=fig, ax=axes[n, m]) # Manual fix to symmetry for theta = 0:2pi
# Polar subplot, with bounds checking
# Polar subplot, with bounds checking
# if (m+1)+((n-1)*sPlots[0])>(sPlots[0]*sPlots[1]):
if (m+1)*(n+1)>(sPlots[0]*sPlots[1]):
pass
elif (n+1)>sPlots[0]:
pass
else:
axes[n,m].plot(np.concatenate((tList, tList+pi)),np.concatenate((Ytp, Ytp)))
axes[n,m].set_title('t = {:3.0f} ps'.format(tIn[tPlot]))
axes[n,m].set_rticks([0.1, 0.2]) # Reduce radial ticks
# Subplot indexing
m += 1
if m >= sPlots[1]:
m = 0
n += 1
# plt.subplots_adjust(hspace=0.5) # Fix overlapping titles
fig.tight_layout() # Or use tight_layout.
plt.show()
At various t...
/tmp/ipykernel_2590/3300663484.py:20: DeprecationWarning: `np.int` is a deprecated alias for the builtin `int`. To silence this warning, use `int` by itself. Doing this will not modify any behavior and is safe. When replacing `np.int`, you may wish to use e.g. `np.int64` or `np.int32` to specify the precision. If you wish to review your current use, check the release note link for additional information.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
Atp += angMomTerm*Ynm(np.int(K),np.int(Q),theta,phi) * A[row]/norm # Add TKQ*Y(K,Q) term
4.1.5. Versions#
import scooby
scooby.Report(additional=['qbanalysis','pemtk','epsproc', 'holoviews', 'hvplot', 'xarray', 'matplotlib', 'bokeh'])
| Tue Jun 03 16:53:17 2025 UTC | |||||||
| OS | Linux (Ubuntu 24.04) | CPU(s) | 4 | Machine | x86_64 | Architecture | 64bit |
| RAM | 15.6 GiB | Environment | Jupyter | File system | ext4 | ||
| Python 3.10.11 (main, Sep 30 2024, 21:36:13) [GCC 13.2.0] | |||||||
| qbanalysis | 0.0.1 | pemtk | Module not found | epsproc | 1.3.2.dev0 | holoviews | 1.20.2 |
| hvplot | 0.11.3 | xarray | 2022.3.0 | matplotlib | 3.5.3 | bokeh | 3.7.3 |
| numpy | 1.23.5 | scipy | 1.15.3 | IPython | 8.37.0 | scooby | 0.10.1 |
# # Check current Git commit for local ePSproc version
# from pathlib import Path
# !git -C {Path(qbanalysis.__file__).parent} branch
# !git -C {Path(qbanalysis.__file__).parent} log --format="%H" -n 1
# # Check current remote commits
# !git ls-remote --heads https://github.com/phockett/qbanalysis
# Check current Git commit for local code version
import qbanalysis
!git -C {Path(qbanalysis.__file__).parent} branch
!git -C {Path(qbanalysis.__file__).parent} log --format="%H" -n 1
* master
4dc763f8848565a5c3d947470fb3eb687d07ba5d
# Check current remote commits
!git ls-remote --heads https://github.com/phockett/Quantum-Beat_Photoelectron-Imaging_Spectroscopy_of_Xe_in_the_VUV
4dc763f8848565a5c3d947470fb3eb687d07ba5d refs/heads/master
2ff23ede221ac1a0ae8b5351c6c505a6ecd1b65d refs/heads/uncertainties